At the time of pushing gordon to GitHub (github.com/aixnr/gordon), I had 35 PDF documents of journal articles and 22 open tabs on my phone, and maybe several more of them as screenshot of title and links on my phone gallery.
I stopped aiming to read them from cover to cover, but to read just enough to understand the questions, the premises, the techniques, and the conclusions.
Over the years since graduate school (2017 - 2022) all the way to postdoc (2022 - 2024), I experimented with ways to archive and annotate papers I have come across.
The most recent iteration of this effort is a .json file containing metadata with tags that summarize/describe the work succintly.
This .json file is then processed into an HTML page:
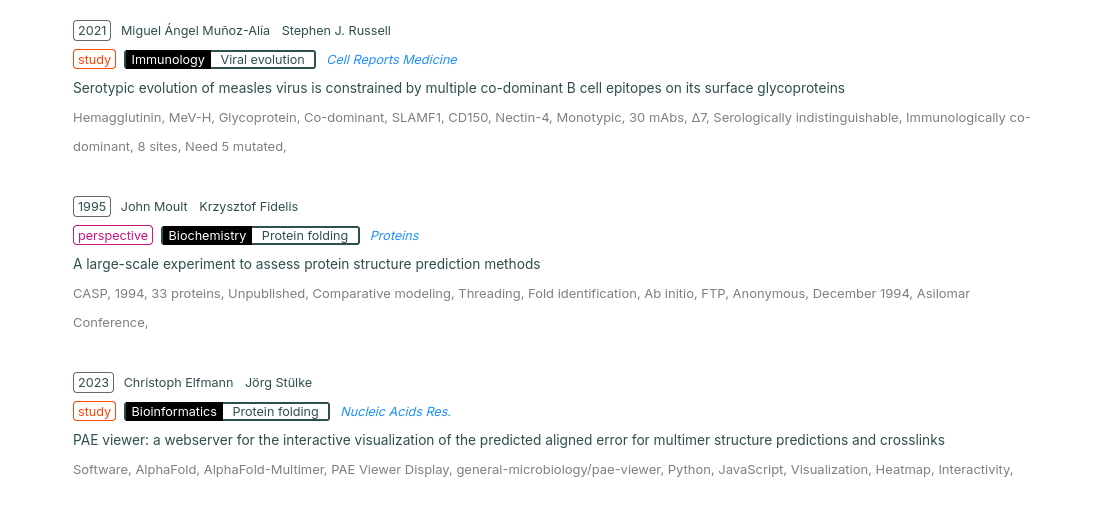
This iteration also includes a fancy hand-rolled search function:
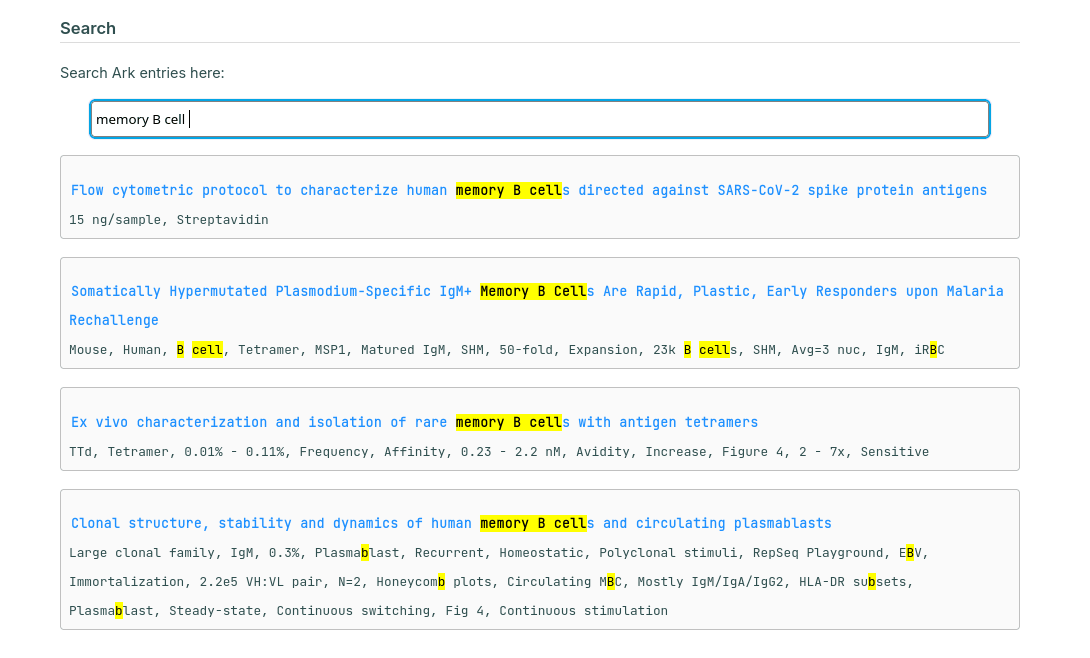
I am currently content with the archival process, much less so with the reading process. Reading takes a lot of work, and often the reward for reading a paper can be marginal. Or, there were times I could be too stupid to understand the implication of certain studies. For studies that I am familiar with the background and the techniques, I could skip most of the fluffs to find exactly what I wanted to learn. This is true for studies in the realm of viral immunology and antibody response, but much less with everything else.
I wrote gordon, a minimal implementation of RAG (Retrieval-Augmented Generation) using the popular LangChain library basically in an afternoon, thanks to 3 large language models (LLMs) that assisted me with the process: GPT-4.1 Mini, GPT-5, and gpt-oss-20b.
Initially this was an experiment to try out the open source gpt-oss-20b released by OpenAI, a model small enough I could run on my AMD Radeon RX 7900 XT (20 GB VRAM) GPU.
This write-up is not meant to be a tutorial, rather just a walkthrough of gordon.
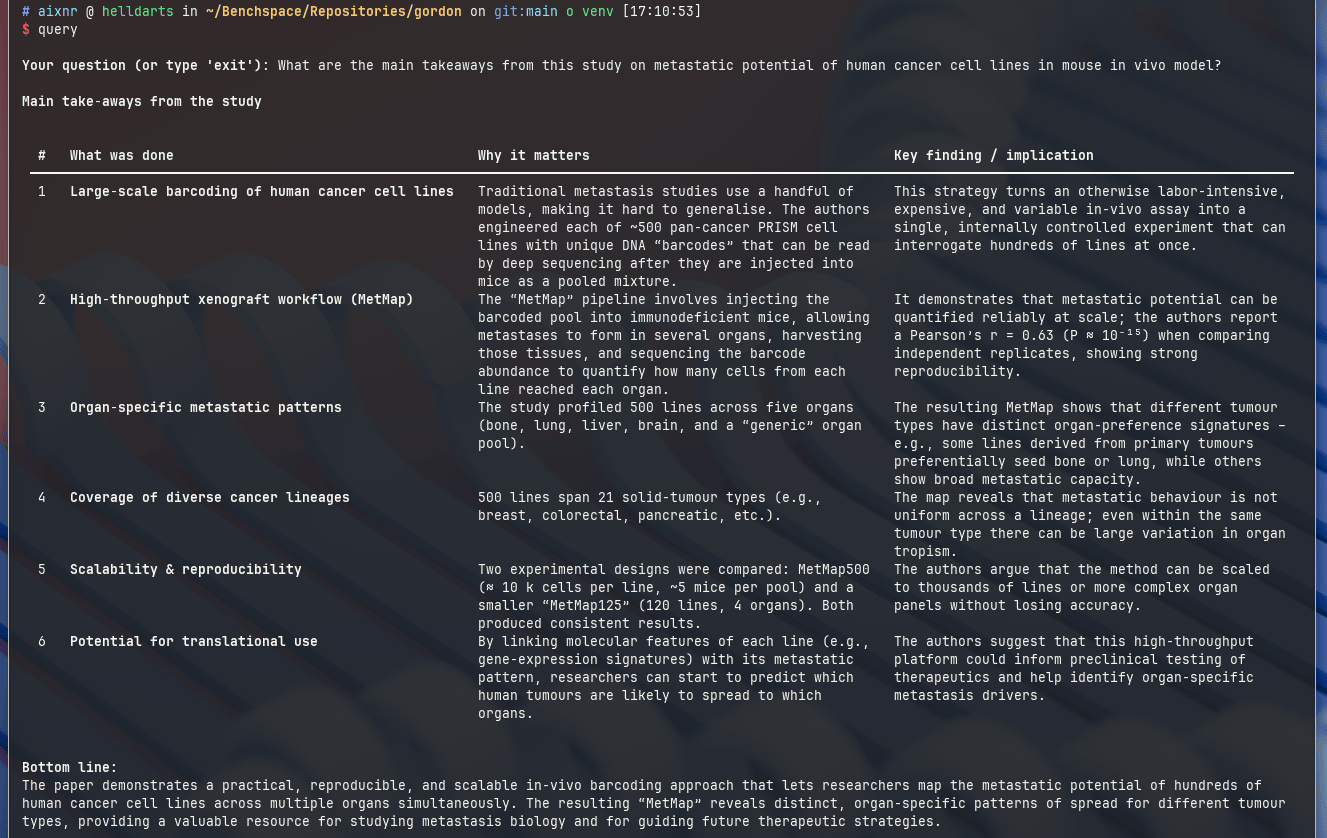
Here are some screenshots from a reading session with a paper by Xin Jin et al. (Nature, 2020): "A metastasis map of human cancer cell lines".

The ingesting process (with the ingest-doc command) for 1 article took 20 seconds to complete.
For this 14.9 MiB (21-pages document), the faiss_index directory containing the indices occupied 583.4 KiB on disk.
With gpt-oss-20b as the chat model (default), it loves producing a table as an output.
While Qwen3-14b likes producing a list:
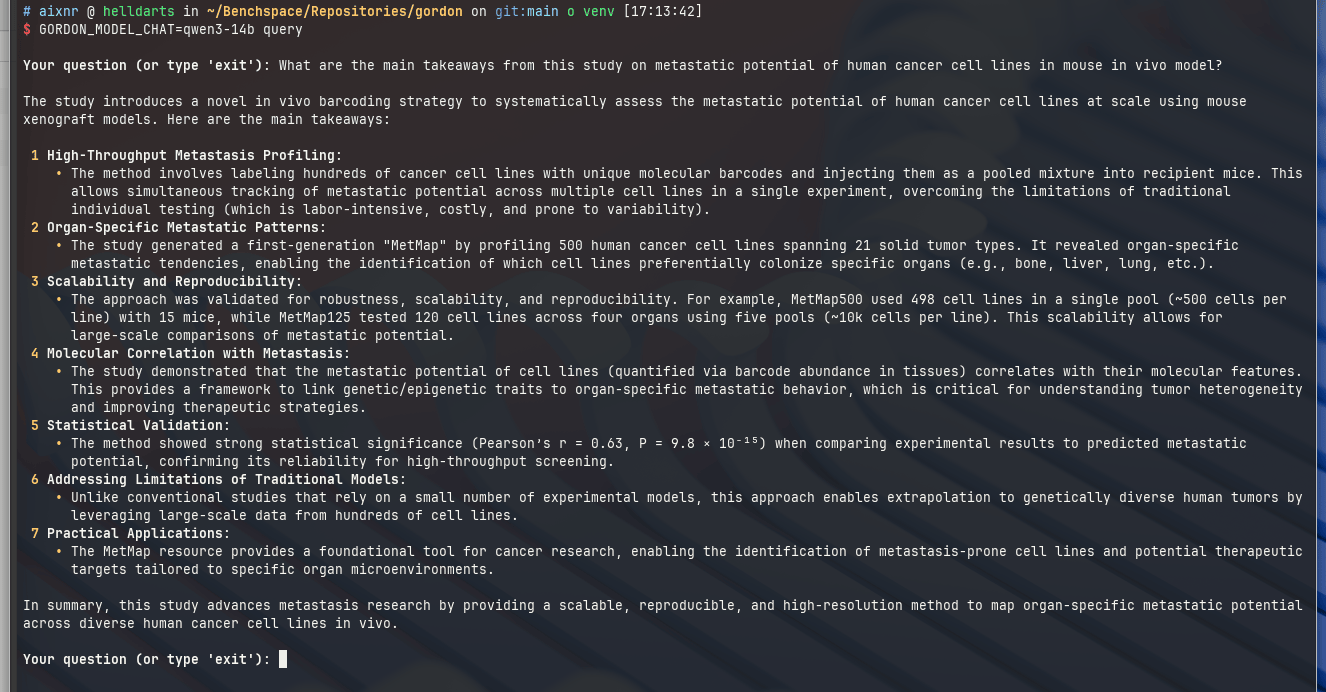
Thanks to the Textualize/rich python library that renders the markdown output to the terminal with proper styling.
Note that user can choose the embedding and chat models by setting the GORDON_MODEL_EMBEDDING and GORDON_MODEL_CHAT environmental variables (env vars).
The OpenAI-compatible API endpoint can also be customized with GORDON_MODEL_ENDPOINT (currently defaults to LM Studio; set the API key with GORDON_API_KEY).
These two examples are clearly contrived, therefore I asked two follow-up questions: (I) List the ranking of organ tropism as described by this study, and (II) Do brain tumor cell lines primarily metastasize to the brain?:
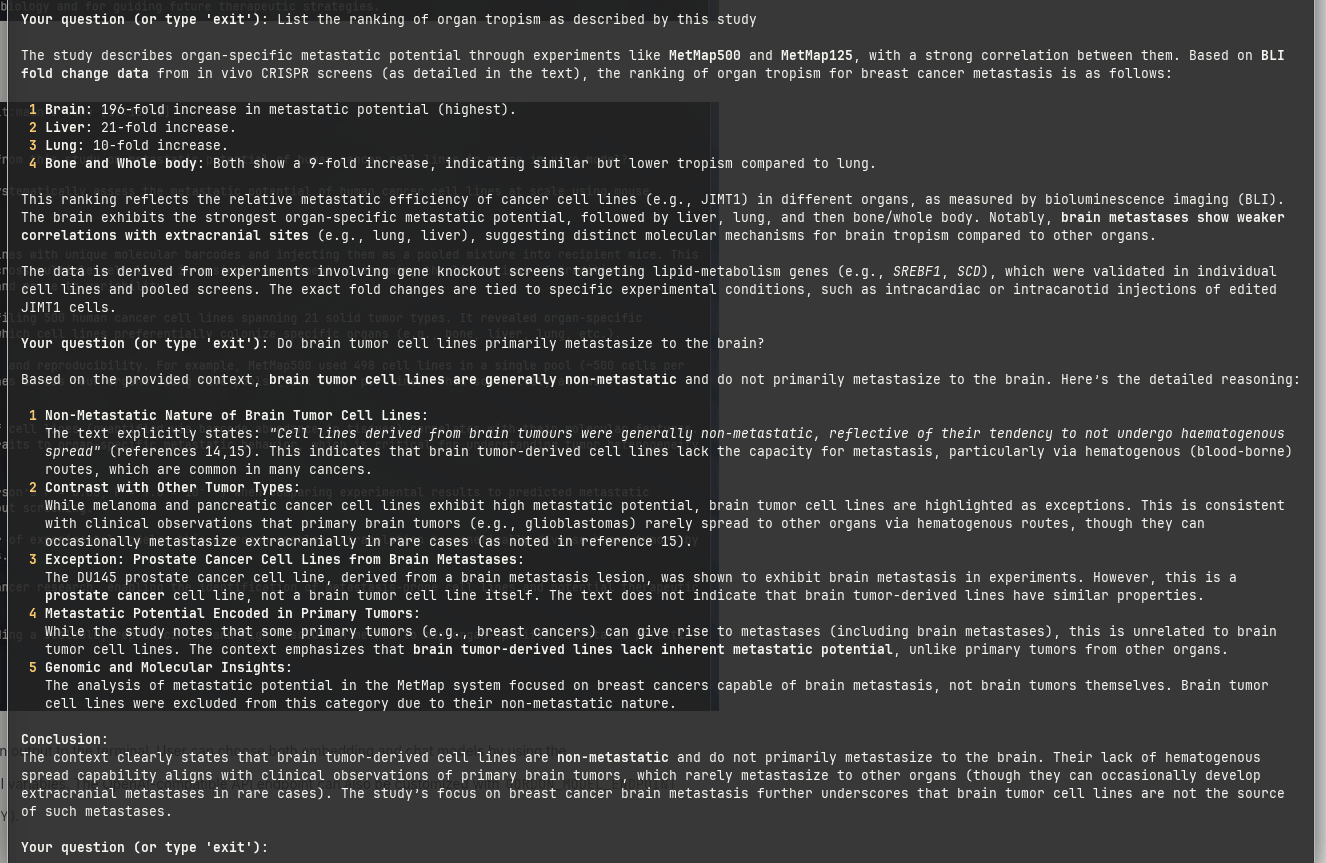
I learned something peculiar about brain tumor cell lines, and a fun fact about the DU145 cell line.
From these simple examples, gordon's utility should now become apparent.
Reading a study involves a linear process (from beginning till end with some dives into the method section).
Published articles nowadays are thick with terminologies, so much so that when we skip the introduction section we could have missed 15-20 definitions.
It becomes terse really quickly.
With gordon, I can get to the points that I care about, and it can me to clarify concepts or information that I missed.
Using this cancer cell line metastasis study as an example, I asked gordon if the authors controlled for synergistic/additive effects of tumor cell lines interacting with each other post-inoculation.
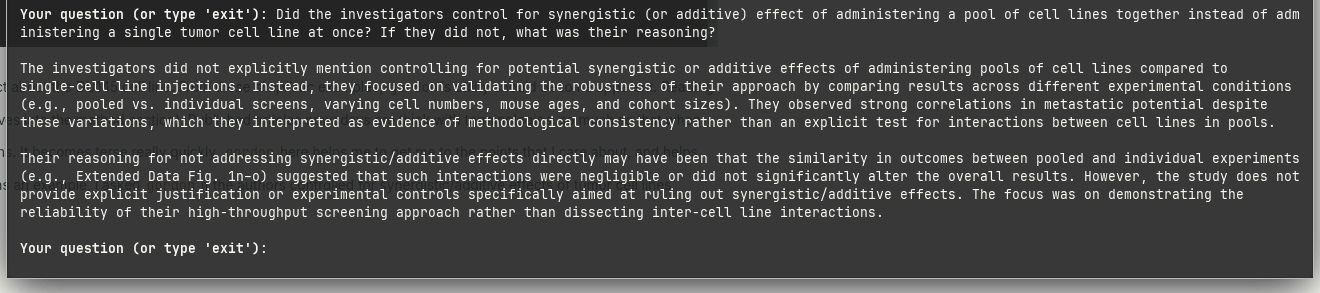
It seems like they did not.
Additionally, when needed, we can instruct gordon to indicate the sources with the --print-context flag.
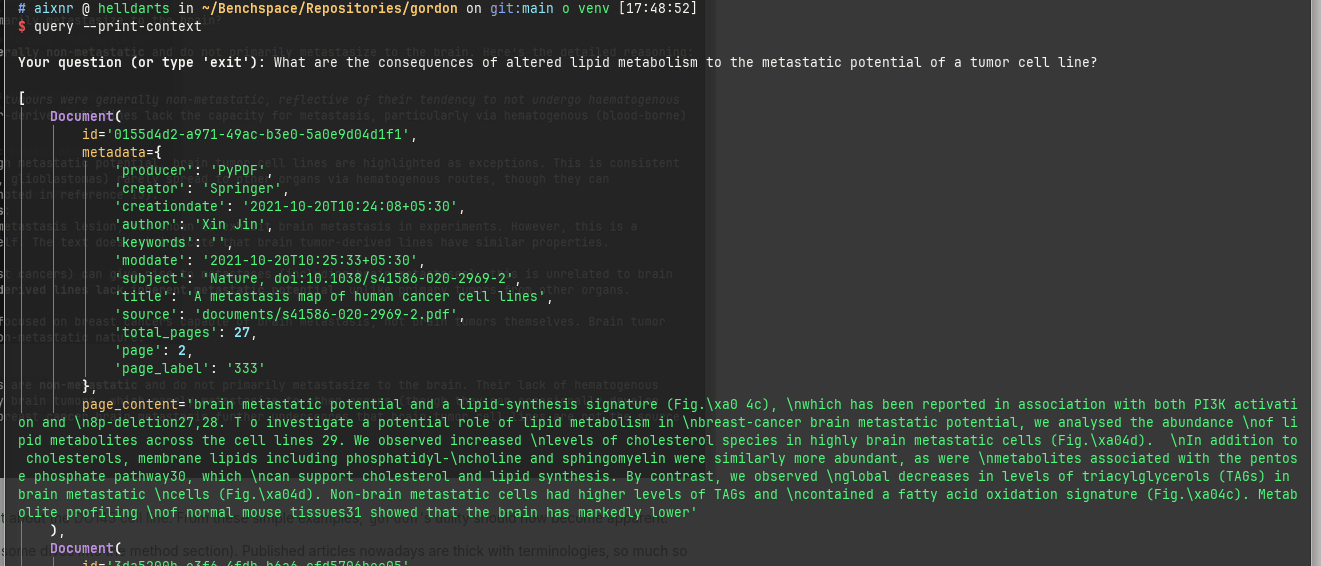
This is all fun and interesting
Building Gordon
As aforementioned, this was an exercise that turned out to be useful. I could have called it a day with Elicit (free tier available, paid tier starts at USD12 monthly) if I wanted a virtual assistant to comb through papers. However, I have a reasonably decent AMD GPU[1] and small LLMs are competent enough for basic tasks. I then propose a set of constraints for this small project.
- Minimal codebase and minimal dependencies.
- 4
.pysource files with a combined 525 lines including comments and whitespaces. - 8 dependencies defined in
pyproject.toml.
- 4
- Somewhat modular:
- I can choose models for embedding and chat at runtime.
- Console scripts
ingest-doc,ingest-web, andqueryhave argument parsers that you can use to customize a handful of options at runtime.
- Operational when offline:
- Tested with connections to LM Studio, but any Llama-based server with OpenAI-compatible API should work.
- Can ingest from PDF documents and from web.
ingest-doclooks for PDF inside thedocumentsubdirectory (default; user-configurable with--doc-dir).ingest-weblooks forweb_sources.json(default; user-configurable by issuingingest-web path/to/file.json).
- Web interface would be nice:
- Currently low priority.
- Maybe later? Maybe a drag-and-drop interface to drop paper and a list interface to input web addresses.
- Can configure several other runtime parameters?
- Probably an interesting future enhancements.
gordonshould not aim to do everything.- It needs to do 1 thing and do it well enough.
- This is just a hobby, it won't be big and professional like Elicit.
Installing Gordon
I recommend using uv for managing python virtual environments (venv).
It is fast and convenient.
To clone and then to install gordon
git https://github.com/aixnr/gordon && cd gordon
uv venv venv
source venv/bin/activate
uv pip install .Alternatively, we can install straight from git:
uv venv venv
source venv/bin/activate
uv pip install "git+https://github.com/aixnr/gordon"Examples in the previous walkthrough looked at ingesting PDF documents with ingest-doc.
Here we look at ingest-web with the following web_sources.json file:
[
{
"url": [
"https://en.wikipedia.org/wiki/Retrieval-augmented_generation"
],
"selectors": [".mw-content-container"],
"tags": ["p"]
}
]Either selectors or tags can be defined, but we can also include both at the same time.
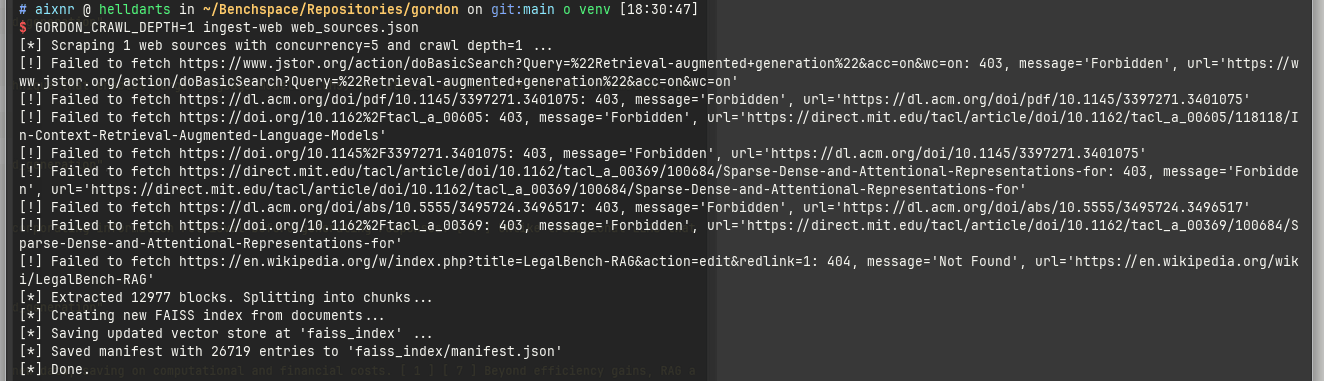
Here we can see ingest-web in action.
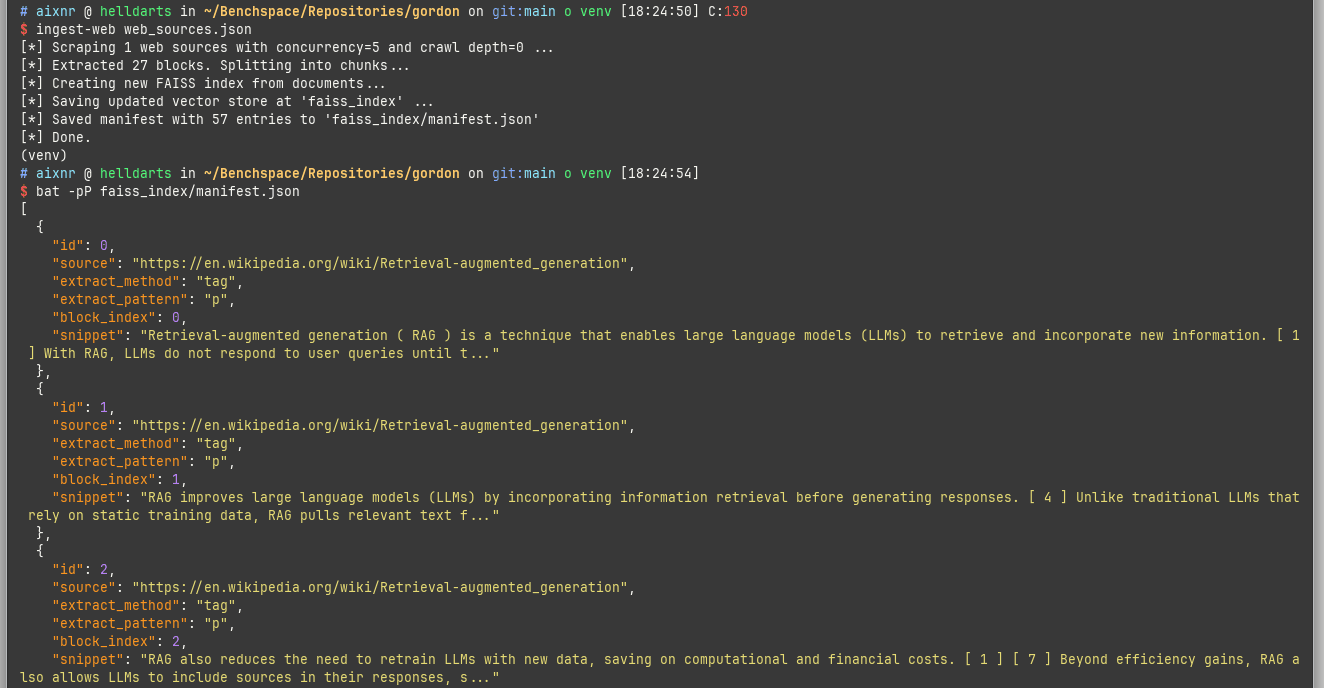
The scraped content can be inspected by viewing the faiss_index/manifest.json file.
I used bat -pP (-p for --plain, -P for --paging=never) to open the file at the terminal as shown above.
ingest-web can also follow links.
By default it does not and we can override this by setting the GORDON_CRAWL_DEPTH env var.
Tip
gordon appends to the same faiss_index if already existed instead of overwriting. This is the intended behavior.
Be aware that crawling to certain addresses may fail for whatever reason.
Following links on a Wikipedia entry may not have been the smartest move; that maneuver took me 11 minutes as it had to comb through 12,977 blocks from a manifest.json file with 213,754 lines with the size on disk of 10.1 MiB.
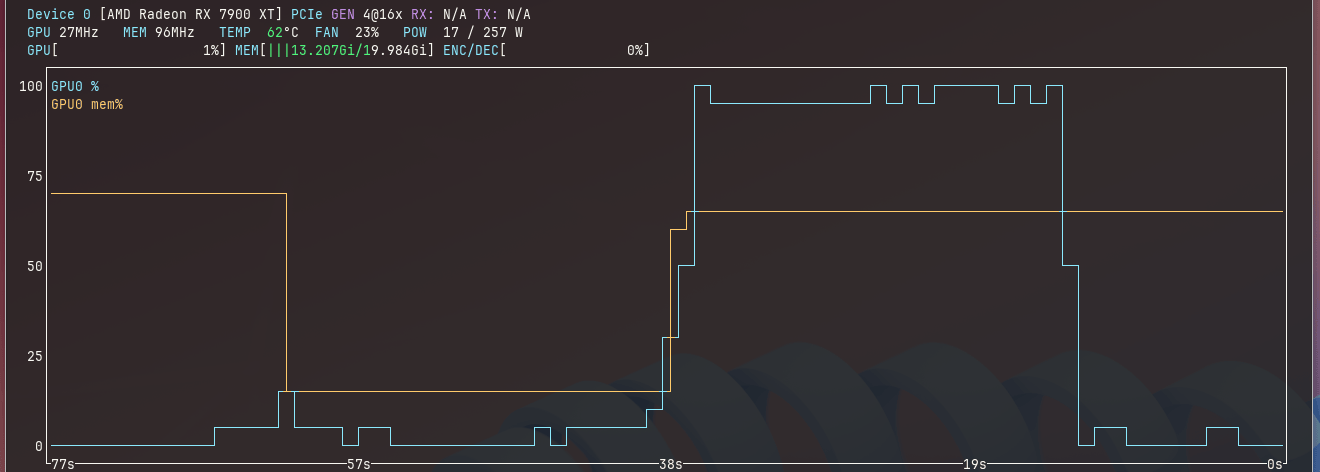
When the models are being actively accessed, GPU utilization is usually maxed out (see the blue line below; output from Syllo/nvtop).
One Notable Issue
Early in the development (first 30 minutes), I learned that OpenRouter only serve chat models but no embedding models. Most tutorials use proprietary OpenAI embedding models for the embedding step. This was the point where I switched to leaning towards LM Studio where at first I wanted to use it with OpenRouter for greater versatility.
The langchain_huggingface.HuggingFaceEmbeddings class (which calls sentence-transformers) worked at the cost of longer duration of the embedding step because I do not have a CUDA device.
# this worked
from langchain_huggingface import HuggingFaceEmbeddings
# model downloaded to
# $HOME/.cache/huggingface/hub
# requires torch and sentence-transformers to be present
# dramatically increased the size of venv to 6.9 GiB
embeddings = HuggingFaceEmbeddings(
model_name="mixedbread-ai/mxbai-embed-large-v1",
model_kwargs={'device': 'cpu'}
encode_kwargs = {'normalize_embeddings': False}
)For reason I did not understand, langchain_openai.embeddings.base.OpenAIEmbeddings did not work with LM Studio.
from langchain_openai import OpenAIEmbeddings
# this does not work
embeddings = OpenAIEmbeddings(
model="mixedbread-ai/mxbai-embed-large-v1",
base_url="http://192.168.1.233:1234/v1",
api_key=SecretStr("dummy-key")
)Both LM Studio and ingest-doc reported the same error: 'input' field must be a string or an array of strings.
Testing the endpoint with curl, however, worked.
curl http://192.168.1.233:1234/v1/embeddings \
-H "Content-Type: application/json" \
-d '{
"input": "the quick brown fox jumps over the lazy dog",
"model": "mixedbread-ai/mxbai-embed-large-v1"
}'
## LM Studio returned the following to the console
# 2025-08-10 20:21:00 [INFO] Received request to embed: the quick brown fox jumps over the lazy dog
# 2025-08-10 20:21:00 [INFO] Returning embeddings (not shown in logs)After consulting with GPT-5, we arrived to this custom class LocalOpenAIEmbeddings(Embeddings) that worked.
class LocalOpenAIEmbeddings(Embeddings):
def __init__(self, model, base_url=api_endpoint, api_key=api_key):
self.model = model
self.base_url = base_url
self.api_key = api_key
def embed_documents(self, texts):
results = []
for text in texts:
resp = requests.post(
f"{self.base_url}/embeddings",
headers={
"Content-Type": "application/json",
"Authorization": f"Bearer {self.api_key}"
},
json={"model": self.model, "input": text} # single string
)
resp.raise_for_status()
results.append(resp.json()["data"][0]["embedding"])
return results
def embed_query(self, text):
return self.embed_documents([text])[0]
embeddings = LocalOpenAIEmbeddings(model=model_embedding)While, sadly, Nvidia is more prevalent in this space because everything is optimized for CUDA. ↩︎
Published on 2025/Aug/11 // Aizan Fahri
aixnr[at]outlook[dot]my